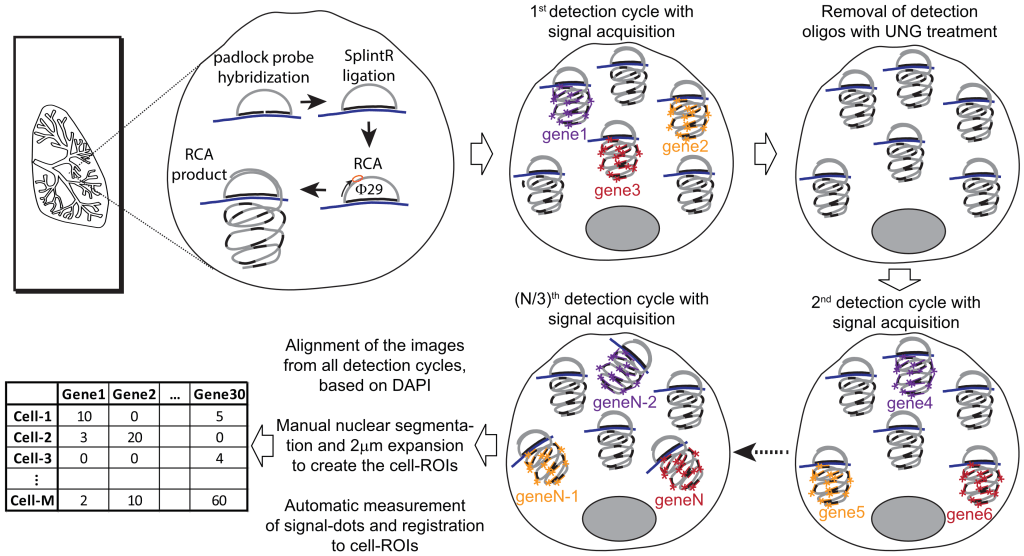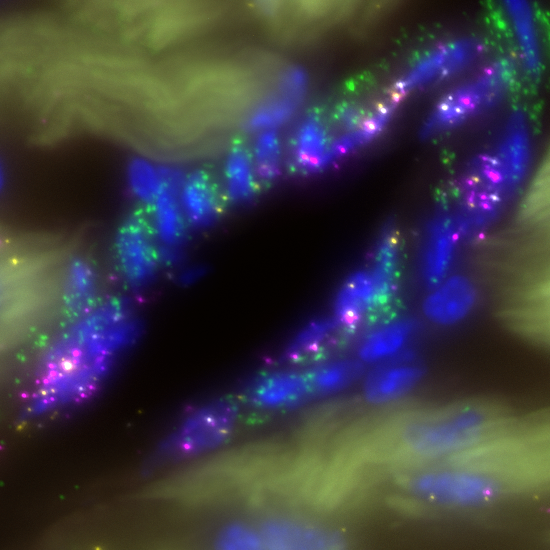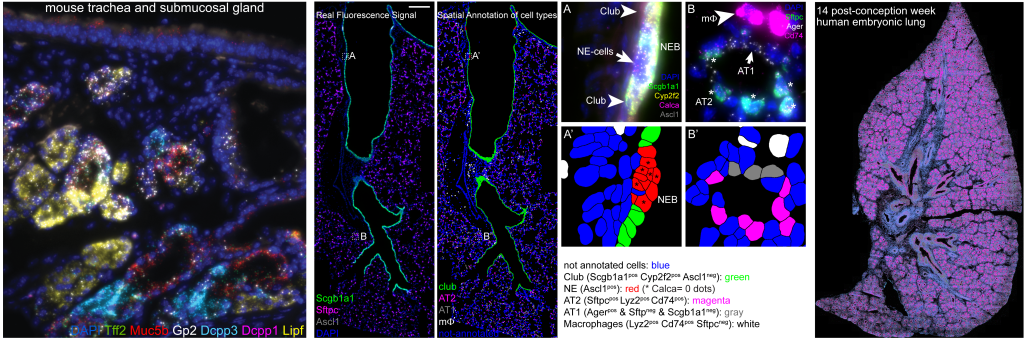
SCRINSHOT (Single-Cell Resolution IN Situ Hybridization On Tissues) is a sensitive, multiplex RNA mapping approach. It utilizes direct hybridization of padlock probes on mRNA, followed by circularization with SplintR ligase and rolling circle amplification (RCA) of the hybridized padlock probes. The sequential detection of RCA-products using fluorophore-labeled oligonucleotides profiles allows the mapping of 50-70 different RNA species (mRNA, log non-coding RNAs) in thousands of cells in tissue sections. The amenability, multiplexity and quantitative qualities of SCRINSHOT can facilitate single-cell mRNA profiling in fresh-frozen and PFA-fixed tissue sections.